Consent
Read carefully and agree on the Terms and Conditions for this challenge, as well as the CAP Data Use Agreement policy document.
In 2011, we held a myocardial segmentation challenge at STACOM 2011 workshop. This challenge was aimed to establish a set of ground truth or consensus segmentation derived from participants. In total there were 100 test cases and 100 training cases randomly selected from the DETERMINE cohort. Three semi- and two fully-automated segmentation algorithms participated in this challenge. The consensus contours were generated based on the Expectation-Maximization principle implemented by the STAPLE method.
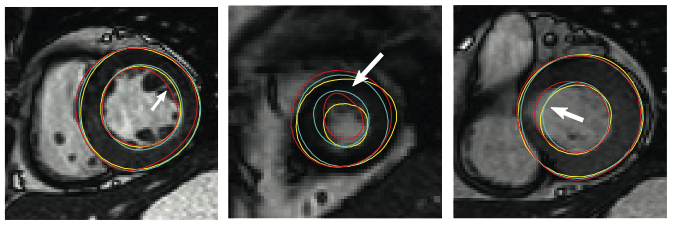
You can read more about this project from [Suinesiaputra et al., 2014].

All challenge data are available for you to download and develop your segmentation method. However, we expect you to return your segmentation results from the test data set so that we can improve our consensus contours. In return, we will perform validation on your segmentation results based on the current consensus contours.
Data description:
| Training data size | 100 |
| Test data size | 100 |
| Contours | Binary mask images (only for training data) |
| Images | Steady-State Free Precession MRI in short and long axis views |
| Pathology | myocardial infarction |
Read carefully and agree on the Terms and Conditions for this challenge, as well as the CAP Data Use Agreement policy document.
Complete and submit the request form below
After approval, you can get a login access to download the data and start developing your segmentation method. Note that you must use the data only for the purpose of this segmentation challenge.
Segment all cardiac frames short-axis MRI views from the test data set. Then submit your segmentation results to us as binary masks, similar with the training mask images.
You will get results back in terms of performance metrics and you will also be able to download the consensus images from the validation dataset.
For submission, please contact us by email.